Radare2 and bioinformatics: a good match?
August 31, 2018
Intro
Ahead of this years’ radarecon, pancake nudged me into discussion we both have about how software reverse engineering and bioinformatics compare and might complement each other, if at all. Inspired by Bunnie Huang’s writeups on (computational) biology as a living example of a cross-domain polymath, I’ll attempt to write down some thoughts and pointers on how radare could be used (or not) in bioinformatics and hopefully manage expectations on what’s possible today.
For starters, back in 2015, the simple Illumina BCL file format got included in radare-extras. As I was providing some specs and explaning how DNA sequencing worked in general, pancake quickly put together a radare plugin for this fairly straightforward file format.
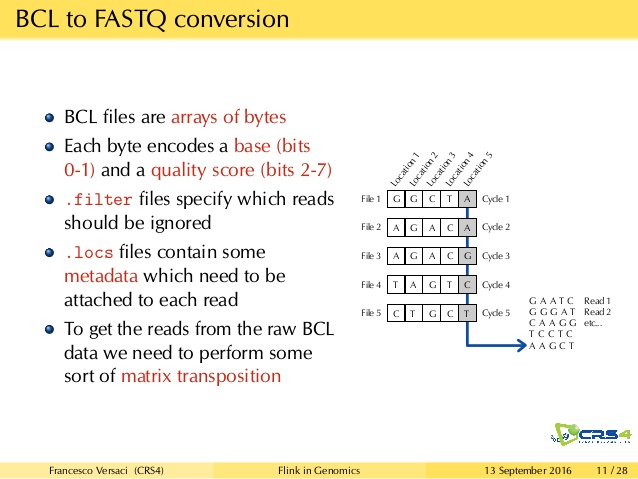
Then fast forward into 2018, radare seems to want more. Here comes the crux of the matter:
Since DNA and ASM are just code running on different architectures, we can reuse the same radare2 reversing workflow right? RIGHT?
Are they?
Before jumping into sleepless nights of unstoppable implementation, please take the time to read two fun papers touching both domains of reverse engineering and biology. They reveal how different (or similar) electronics manufacturing and biology can be.
As stated in biologists trying to figure out a radio paper:
“(…) the commonality of the language allows engineers to identify familiar patterns or modules (a trigger, an amplifier, etc.) in a diagram of an unfamiliar device”
Food for thought: how “unfamiliar” of a “device” is biology itself versus human-manufactured consumer wares?
In another, more recent, neuroscinece paper, “Could a Neuroscientist Understand a Microprocessor?", some insights come up:
Much has been written about the differences between computation in silico and computation in vivo (…) the stochasticity, redundancy, and robustness present in biological systems seems dramatically different from that of a microprocessor. But there are many parallels we can draw between the two types of systems.
Bottom line is, they are definitely different yet similar in some instances. Without getting overwhelmed by the huge, sometimes messy, amount of domain-specific knowledge to digest, how can we score “quick wins” for radare2 if an implementation under radare-extras starts to happen?
Observing bioinformatics from the RSE perspective, I see great contributions that could happen in three areas:
- EBA: Exploratory Bioinformatics Analysis.
- Scientific algorithm optimization and security.
- Outreach.
Radare2 TL;DR for bioinformaticians
Here’s some explanation on how radare works from a user perspective. If you barely recall what assembly was from school, I’ll leave you in good hands to catch up with ARM assembly here
Also, some relatively recent UI eyecandy from Cutter:

Often touted as “steep learning curve” framework due to its commands, radare2 has been misunderstood for years, since in reality, keybindings allow for distraction-free fast iteration during binary analysis.
Bioinfomatics TL;DR for radare2 developers
If you are a r2 developer, those are the formats radare would need to understand and implement to be minimally interesting for our biologist neighbors (optional ones, inside parenthesis):
Now, one could go the hardcore pancake/Feynman (brentp?) way and implement file parsers from scratch or use some third party library such as htslib.
After basic read/write functionality is in place, I think that a potential first win would be to have the “Midnight Commander”-equivalent of radare4bio for curious and impatient bioinformaticians.
There’s great educational potential if this is implemented right since radare allows for fast VIM-like iteration and speed during complex analysis.
For instance, being able to examine individual reads with VIM shortcuts, flip/cycle CIGAR encodings, like with the radare2 bit editor:
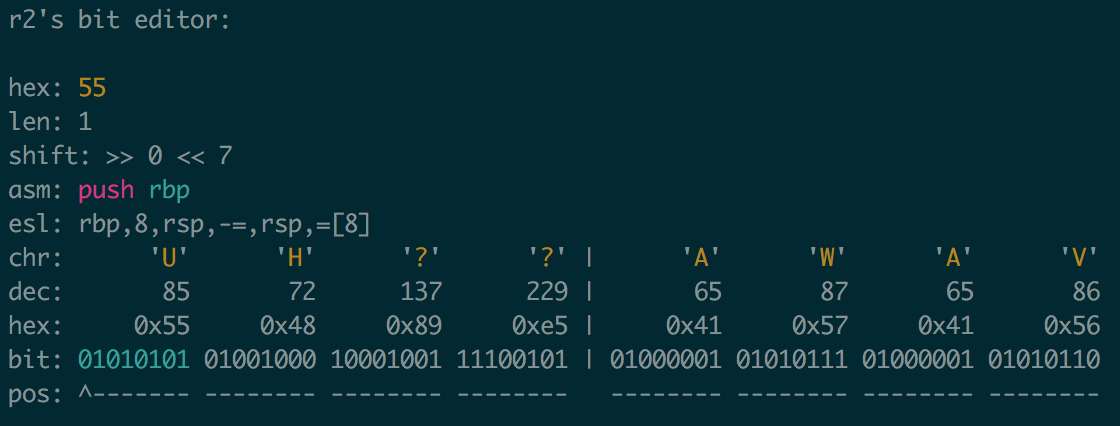
Group reads by some arbitrary criteria, subsample, filter them, write out, etc…
That is, FAST exploratory bioinformatics analysis (EBA) without the overhead of writing discrete commands or putting together workflows, pipelines and/or lengthy documentation
Outro
-
How would radare really help with “biology reverse engineering”?
-
How can radare absorb those “extras” without introducing a vast dependency tree of bioinfo software? Perhaps a clean-room implementation is still of interest nowadays?
-
Would all that coding effort be worth it?
Those are open questions at the time of writing this, but here are some opportunities:
When bioinformaticists analyze data (and are not waiting for big computations to complete), it is hugely helpful to iterate fast on a particular question. Keeping focus on the task at hand while answering questions fast and accurately is immensely valuable.
Radare2 is well positioned in this regard, allowing for fast adhoc analysis for the reasons stated before (VIM-like blazing speed shortcuts, focus on speed).
As a former colleague pointed out, Bioformatics (and scientific software in general) is in dire need for optimization and good software engineering at several levels: Storage, data processing, security (read this!), good software design patterns, etc…
If all else fails, the outreach value of getting reverse engineers poking into computational biology is in itself, a huge win, IMHO.
If you are still reading this, I’m honored :)
Please ping me physically during the radare2 2018 con or via twitter @braincode if you want to have a chat about this and other random braindumplings.